The book for my package diagnoser has just been published online! You may view it as a Gitbook (https://rs-diagnoser.netlify.com/) or PDF (https://github.com/robertschnitman/diagnoser/blob/master/docs/diagnoser_handbook.pdf).
I recommend reading it if you are interested in a new perspective on diagnosing your model residuals!
Installing diagnoser
The library diagnoser currently is only installable via GitHub and is contingent on R versions at or above 3.4.2. To install the package, first install devtools so that you may make use of the function install_github, referencing diagnoser by the package creator’s username (“robertschnitman”) followed by “/diagnoser” as shown in the code below:
## Ensure that you are running R 3.4.2 or higher.
## Package Dependencies:
# lazyeval (>= 0.2.1)
# Package Imports:
# ggplot2 (>= 2.2.1),
# gridExtra (>= 2.3),
# scales (>= 0.5.0),
# car (>= 2.1)
# Install library necessary for installing diagnoser.
install.packages("devtools")
# Install diagnoser via devtools.
devtools::install_github("robertschnitman/diagnoser")Example 1: diagnose()
model.lm <- lm(data = mtcars, formula = mpg ~ wt + gear)
diagnose(model.lm,
fit_type = 'response',
residual_type = 'response')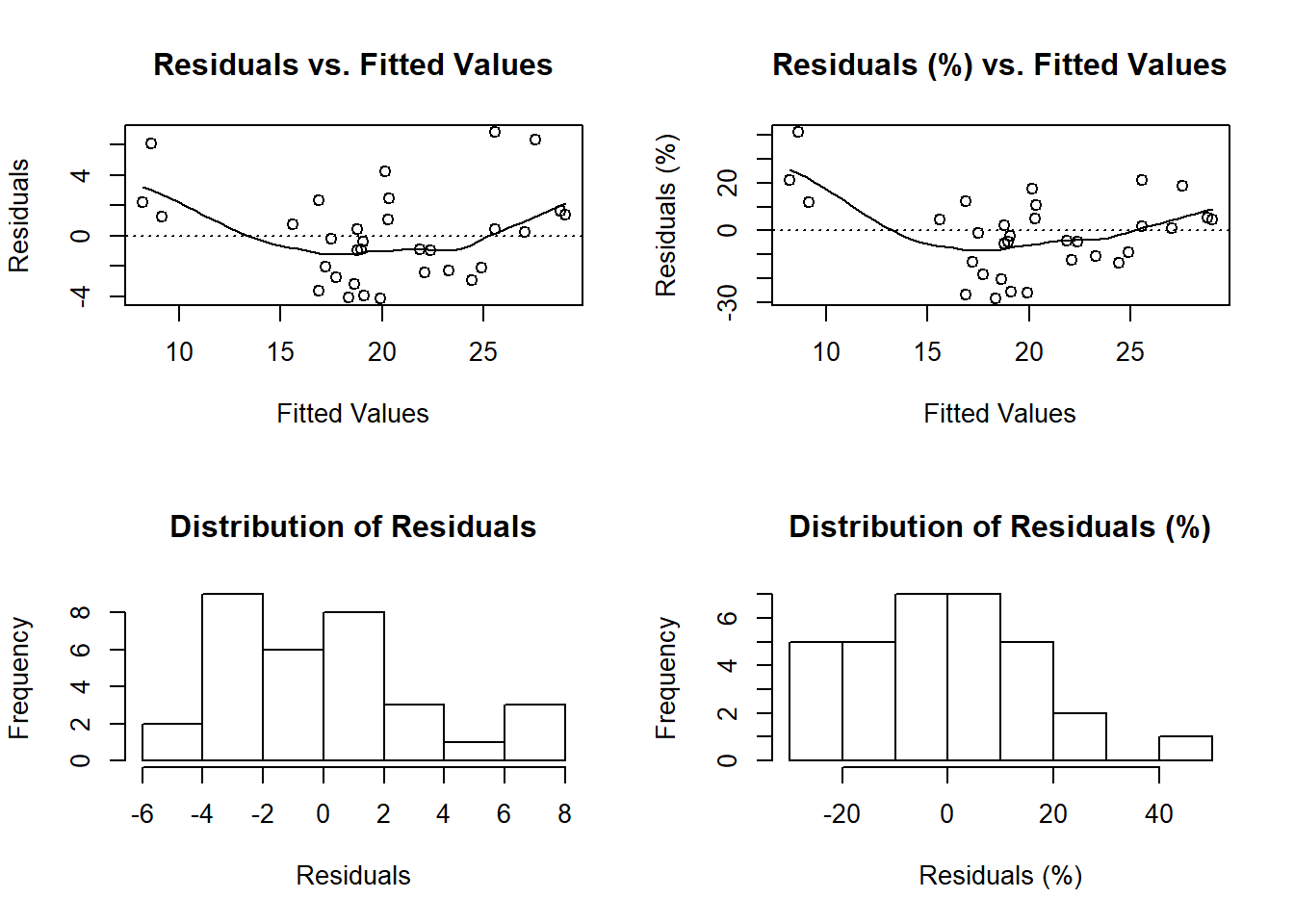
Example 2: ggdiagnose()
ggdiagnose(model.lm,
fit_type = 'response',
residual_type = 'response',
freqpct = TRUE,
alpha = 0.5)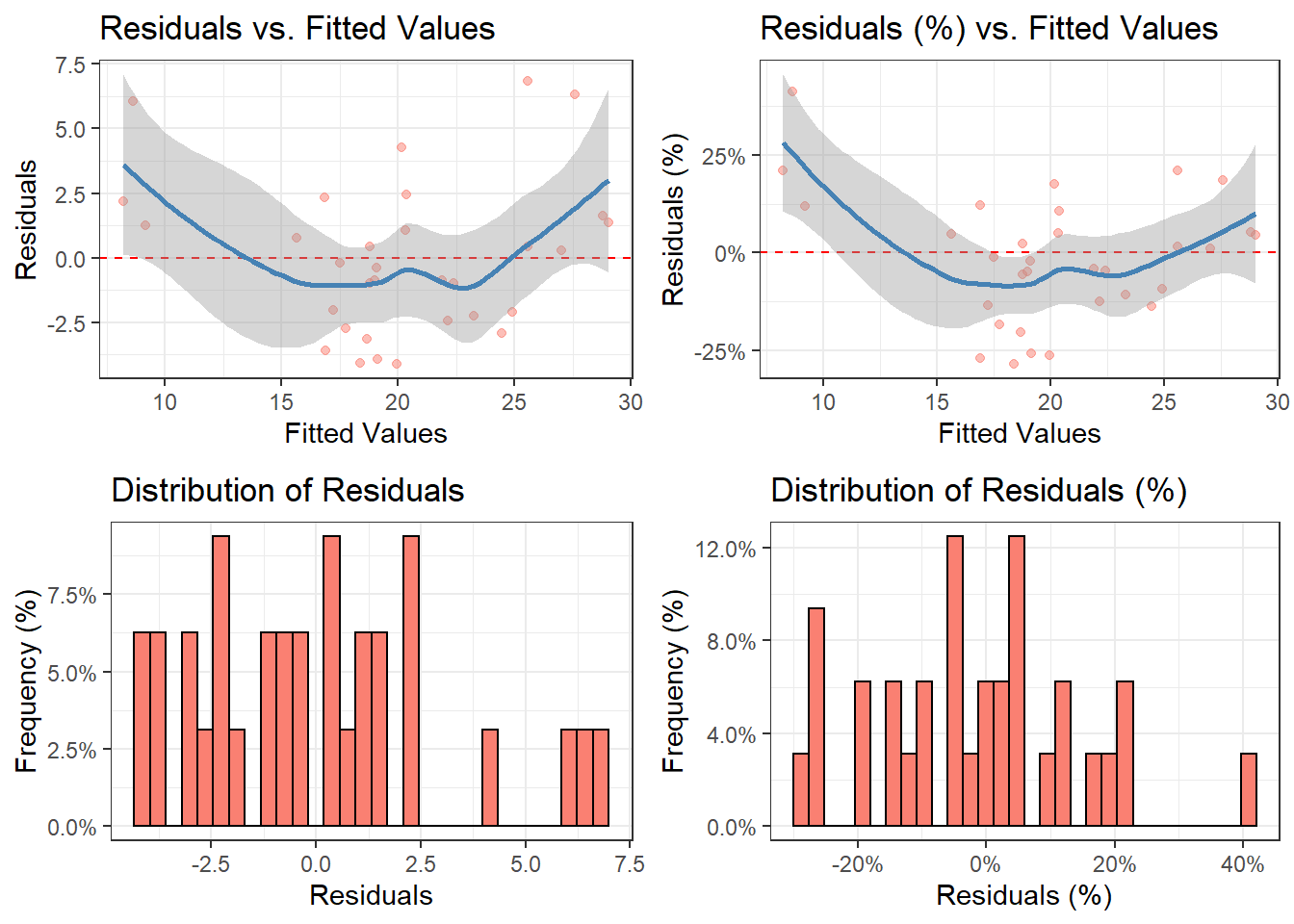
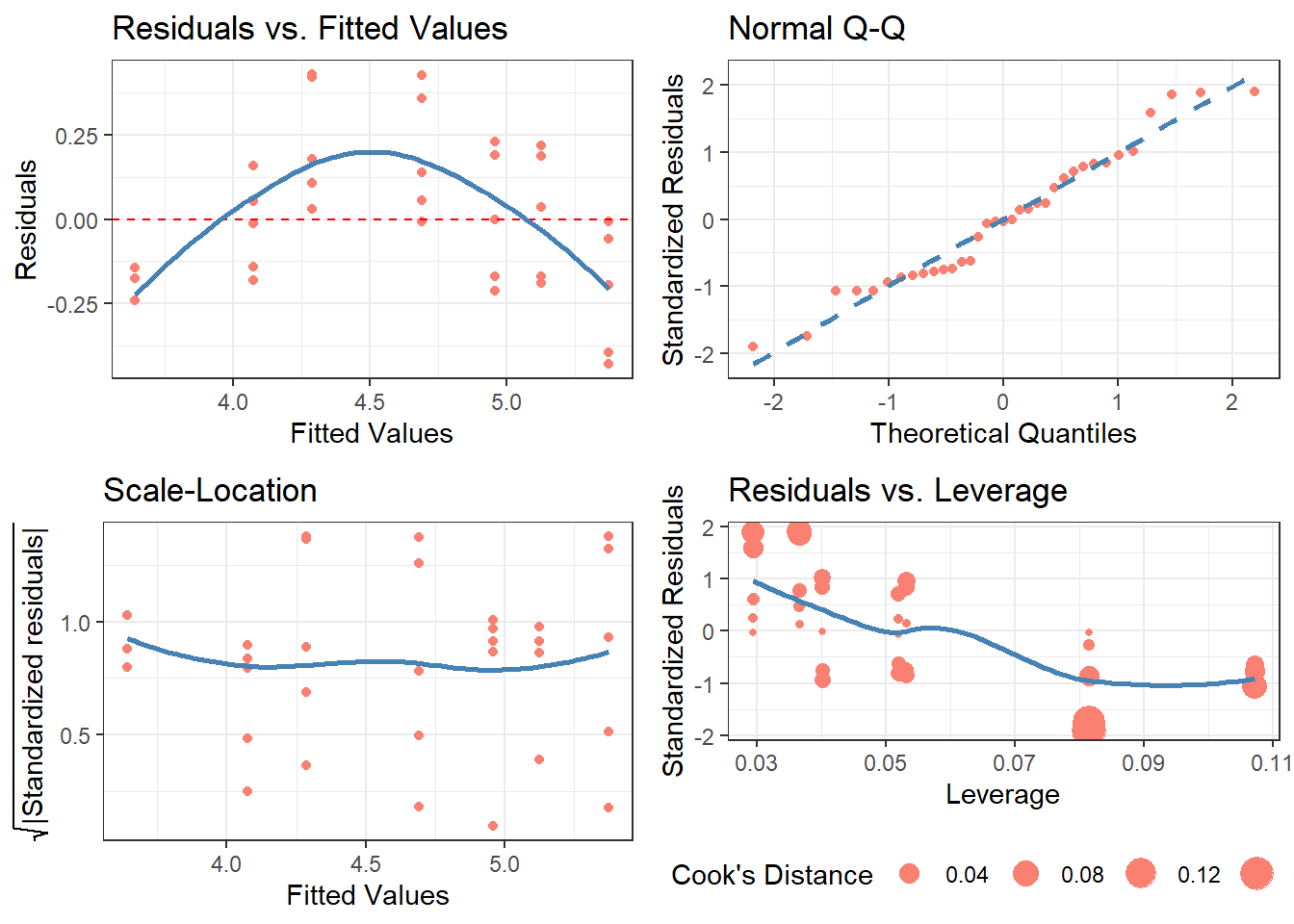
Example 3: cdiagnose()
model.lm <- lm(data = Orange, formula = log(circumference) ~ age)
cdiagnose(model.lm,
fit_type = 'response',
residual_type = 'response',
se = FALSE,
alpha = 1)